Random Fourier Features for Kernel Density Estimation
The NIPS paper Random Fourier Features for Large-scale Kernel Machines, by Rahimi and Recht presents a method for randomized feature mapping where dot products in the transformed feature space approximate (a certain class of) positive definite (p.d.) kernels in the original space.
We know that for any p.d. kernel there exists a deterministic map that has the aforementioned property but it may be infinite dimensional. The paper presents results indicating that with the randomized map we can get away with only a “small” number of features (at least for a classification setting).
Before applying the method to density estimation let us review the relevant section of the paper briefly.
Bochner’s Theorem and Random Fourier Features
Assume that we have data in and a continuous p.d. kernel
defined for every pair of points
. Assume further that the kernel is shift-invariant, i.e.,
and that the kernel is scaled so that
.
The theorem by Bochner states that under the above conditions must be the Fourier transform of a non-negative measure on
. In other words, there exists a probability density function
for
such that
.
where (1) is because is real. Equation (2) says that if we draw a random vector
according to
and form two vectors
and
, then the expected value of
is
.
Therefore, for , if we choose the transformation
with drawn according to
, linear inner products in this transformed space will approximate
.
Gaussian RBF Kernel
The Gaussian radial basis function kernel satisfies all the above conditions and we know that the Fourier transform of the Gaussian is another Gaussian (with the reciprocal variance). Therefore for “linearizing” the Gaussian r.b.f. kernel, we draw samples from a Gaussian distribution for the transformation.
Parzen Window Density Estimation
Given a data set , the the so-called Parzen window probability density estimator is defined as follows
where is often a positive, symmetric, shift-invariant kernel and
is the bandwidth parameter that controls the scale of influence of the data points.
A common kernel that is used for Parzen window density estimation is the Gaussian density. If we make the same choice we can apply our feature transformation to linearize the procedure. We have
where has been absorbed into the kernel variance.
Therefore all we need to do is take the mean of the transformed data points and estimate the pdf at a new point to be (proportional to) the inner product its transformed feature vector with the mean.
Of course since the kernel value is only approximated by the inner product of the random Fourier features we expect that the estimate pdf will differ from a plain unadorned Parzen window estimate. But different how?
Experiments
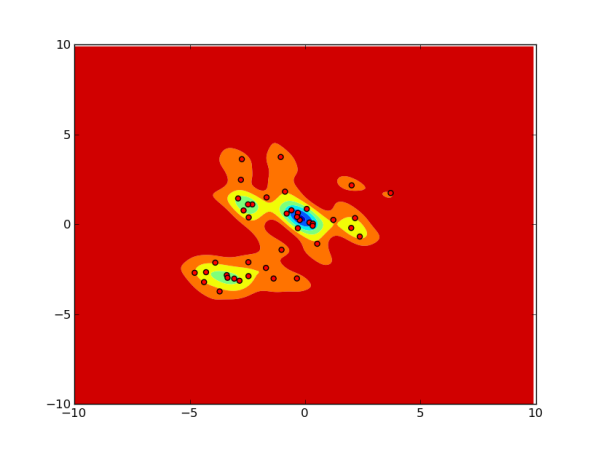
Below are some pictures showing how the method performs on some synthetic data. I generated a few dozen points from a mixture of Gaussians and plotted contours of the estimated pdf for the region around the points. I did this for several choices of and
(the scale parameter for the Gaussian kernel).
First let us check that the method performs as expected for large values of because the kernel value is well approximated by the inner product of the Fourier features. The first 3 pictures are for
for various values of
.
—————————————————————————
—————————————————————————
Now let us see what happens when we decrease . We expect the error in approximating the kernel would lead to obviously erroneous pdf. This is clearly evident for the case of
.
—————————————————————————
—————————————————————————
The following picture for and
is even stranger.
—————————————————————————
—————————————————————————
Discussion
It seems that even for a simple 2D example, we seem to need to compute a very large number of random Fourier features to make the estimated pdf accurate. (For this small example this is very wasteful, since a plain Parzen window estimate would require less memory and computation.)
However, the pictures do indicate that if the approach is to be used for outlier detection (aka novelty detection) from a given data set, we might be able get away with much smaller . That is, even if the estimated pdf has a big error on the entire space, on the points from the data it seems to be reasonably accurate.
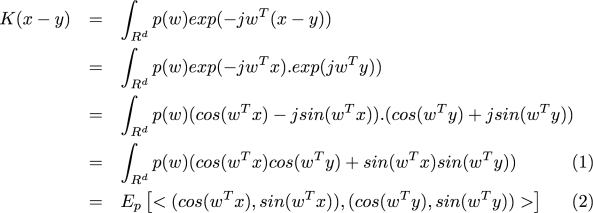






Thank you. I was having some trouble understanding this paper
Thanks a lot it really helps to understand the paper.
thanks a lot. IT would have been great if you can share the code.
Thanks a lot. It was really easy to understand. Can you please upload your codes too.
Thanks